 A paper by UM scholars has been published in Lab on a Chip
A paper by UM scholars has been published in Lab on a Chip
A paper by UM scholars was recently published in Lab on a Chip, a publication under the Royal Society of Chemistry in the United Kingdom. The paper proposes a new technique that can quickly identify the DNA of multiple potential pathogens causing septicemia from a single droplet (1uL). This new technique will allow medical workers to promptly identify pathogens and administer treatment in remote mountainous regions. This is yet another breakthrough in UM’s microchip research. The related system is expected to be put into production within the next two years.
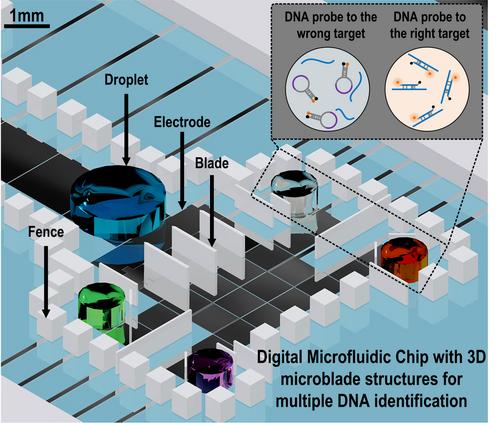
Titled ‘3D Microblade Structure for Precise and Parallel Droplet Splitting on Digital Microfluidic Chips’, the paper was co-written by PhD student Dong Cheng, Prof Jia Yanwei, Dr Gao Jie, Dr Chen Tianlan, Prof Mak Pui-In, Prof Vai Mang-I, and Vice Rector (Research) Prof Rui Martins.
In the study, 3D microblades were innovatively fabricated on the pathway of droplet transportation on DMF chips, providing precise, reversible, volume controllable and parallel droplet splitting. With multiple blades implemented, only two electrodes were needed to simultaneously generate up to four daughter droplets in one step splitting. Using this technique, the DNA of multiple potential pathogens causing septicemia were successfully identified from a single droplet (1uL).
According to Prof Jia Yanwei, analysis of parallel DNA is not difficult; the real challenge lies in the volume of the sample. Traditional PCR machines normally require samples of 25uL, and the analysis can only be done in a laboratory. The new technique developed by UM using digital microfluidics only requires a sample of 1uL to achieve the same results. Moreover, this new technique can be applied to make portable outdoor DNA testing devices.
The main advantage of this system is that it can realise portable DNA testing and diagnose hidden or contagious diseases, which will allow medical workers to promptly identify pathogens and administer treatment in remote mountainous regions.